🎄12 Days of HPC 2022
Graph yourself a merry little Christmas
During the month of December we’re featuring blog posts from researchers from across the University of Leeds showcasing the fantastic work they do using our High Performance Computing system. Follow us @RC_at_Leeds to keep up to date with our 12 days of HPC blog series.
What’s your name?
Lucy Godson
What department do you work in?
School of computing (as a PhD student @ CDT for AI for Medical Diagnosis and Care)
What research question are you trying to answer?
My project looks a melanoma patient data from patients enrolled in the Leeds Melanoma Cohort. Melanoma is the most aggressive form of skin cancer and the fifth most common cancer in the UK. While most of our understanding of patient prognosis focuses on looking exclusively at histopathology images - high resolution images of the cells of a patient’s tumour, recent studies have shown how genetic signatures from a patient’s tumour can also be important in predicting patient outcome. In my research, I am aiming to use artificial intelligence methods to bring this imaging and molecular data together with clinical data, to see if this helps improve our understanding of the disease and patient outcomes.
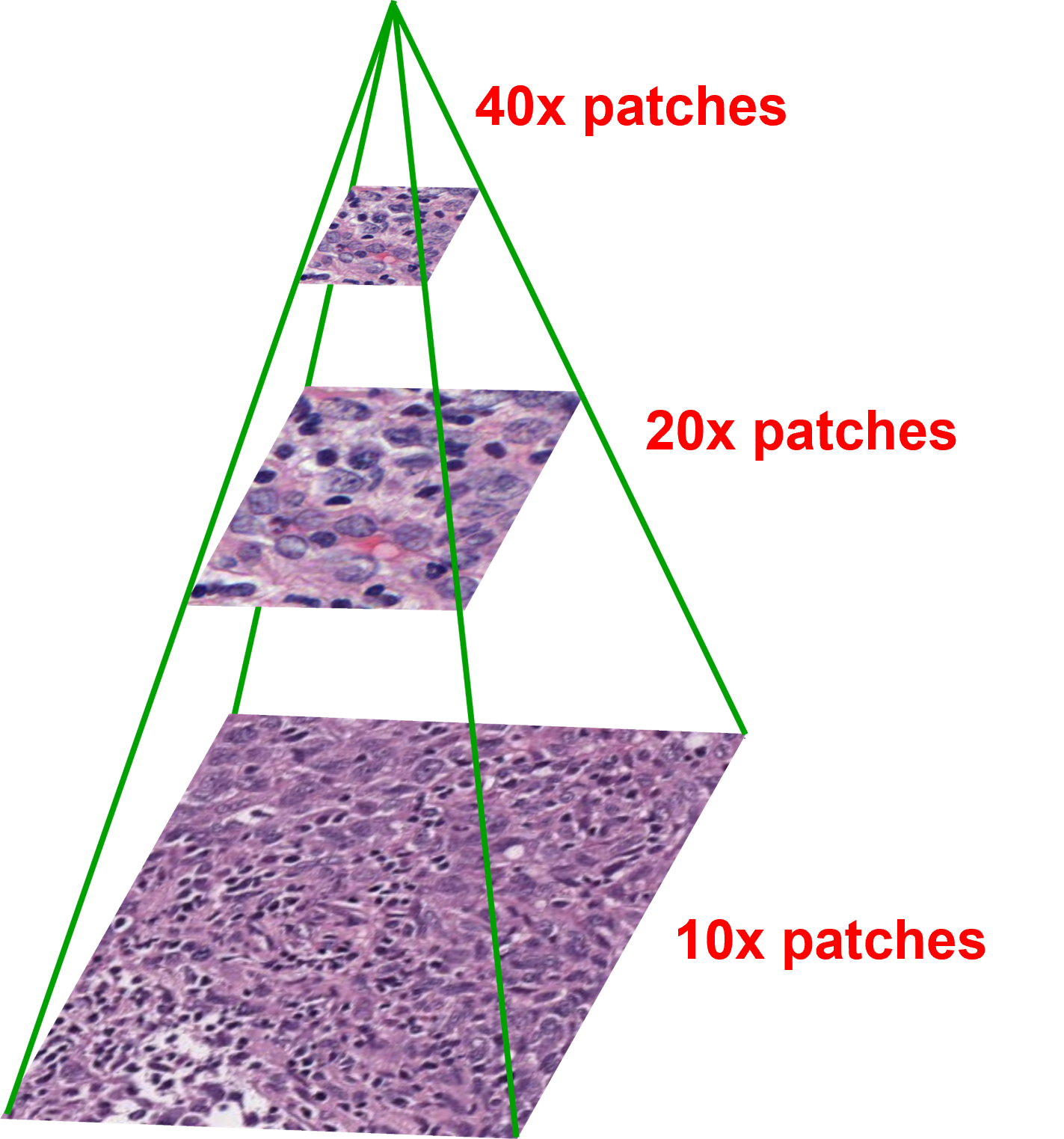
However, histopathology images are large gigapixel images, therefore, it is computationally unfeasible to analyse the whole image at once. Moreover, these images have multiple resolutions to allow pathologists to look at the images at different magnifications for different levels of cellular detail. So, I use patch-based approaches, which divide the images into individual 256 pixel x 256 pixel patches, then use graphs to connect the patches and show connections across different magnification levels within a single image.
What tools or technologies do you use in your research? (Programming languages, packages, APIs)
I mainly code using python scripts, with PyTorch to implement neural networks and PyTorch geometric to create graphs.
How does HPC help your research?
HPC allows me to process and analyse enormous histopathology images (billions of pixels!), using state of the art neural networks. Also the HPC software engineering team have really helped me with setting up environments to run my experiments - package dependency hell is real!
What is the potential impact of your research?
Melanoma is the most deadly form of skin cancer, so being able to better understand the tumour landscape and group patients accordingly, will help patients receive the most beneficial treatments. Hopefully my research is a stepping stone to developing a clinical tool that could help with this.
In your personal opinion what’s the coolest thing about your research?
I really enjoy working in an interdisciplinary team, with clinicians, computer scientists, biologists and research software engineers!
Postscript
I would like to thank the participants of the Leeds Melanoma Cohort for their participation and generosity in this research.