🎄12 Days of HPC
Brain Cancer Code Crackers
During the month of December we’re featuring blog posts from researchers from across the University of Leeds showcasing the fantastic work they do using our High Performance Computing system. Follow us @RC_at_Leeds to keep up to date with our 12 days of HPC blog series.
What’s your name?
Lucy Stead
What department do you work in?
Leeds Institute of Medical Research
What research question are you trying to answer?
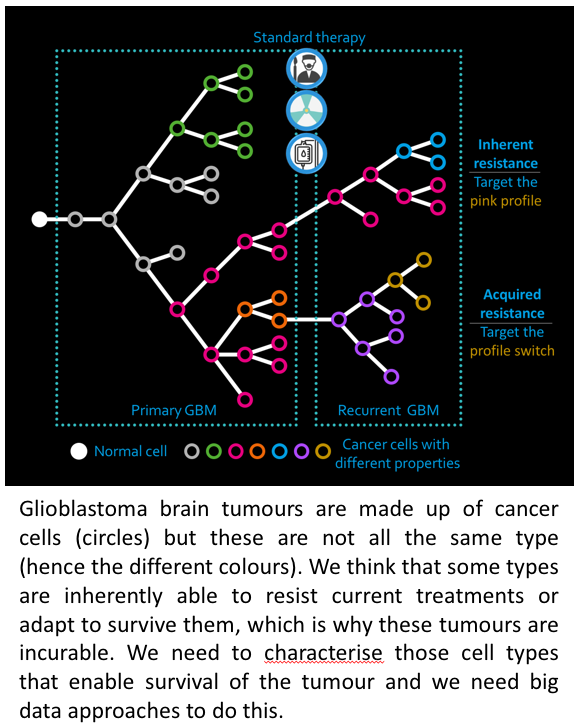
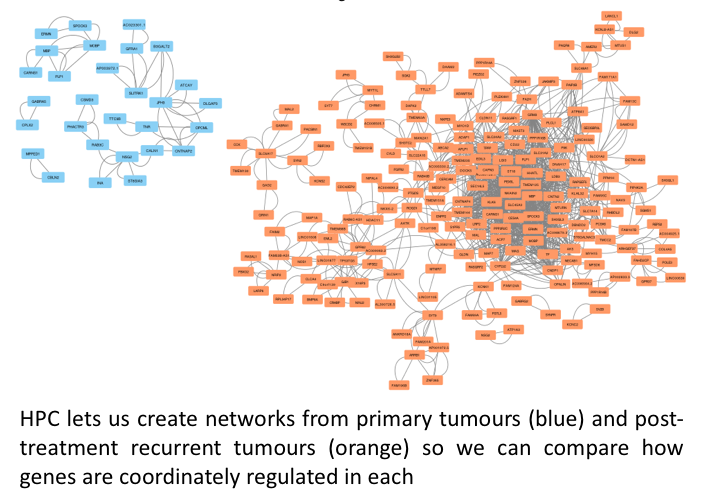
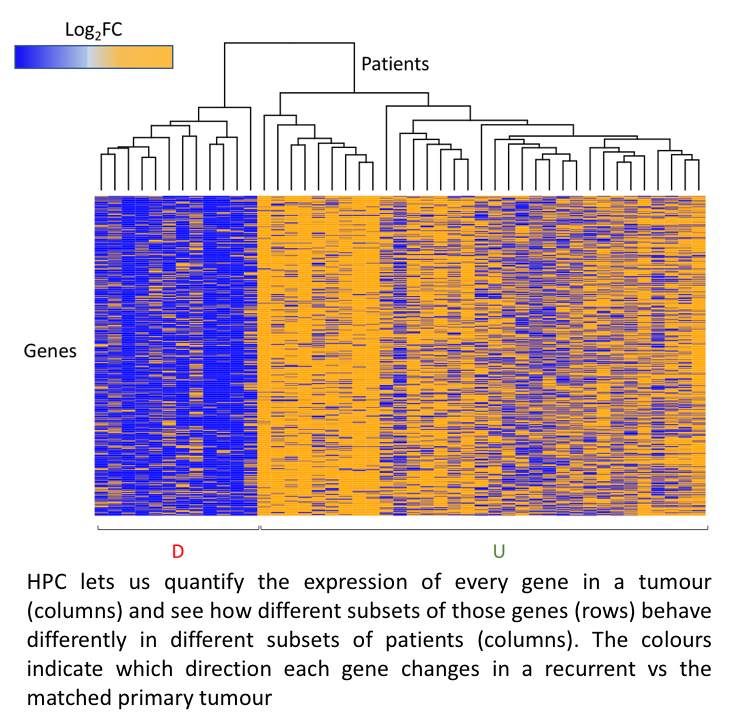
Glioblastoma (GBM) brain cancer is arguably the worst cancer diagnosis a person can receive: it is a deadly and incurable brain cancer with a median survival of just 14 months. GBM tumours infiltrate the surrounding brain, making complete surgical removal impossible. The cells that remain are treatment with both radio- and chemotherapy but some inevitably survive and the tumours grown back in almost 100% of cases. We must understand why GBM cells resist treatment before we can hope to prolong patient survival. Our approach is to collect and compare the molecular profiles (DNA and the information on the genes it encodes) of primary and recurrent GBM tumours to specifically characterise the cells that survived treatment, with the ultimate aim of finding ways to more effectively kill them.
We couldn’t do our research without the HPC - but who are we? My group is a hybrid of experimental and computational biologists but all of them make use of the HPC - and they’re the ones that do the really hard work. My all star group members are:
- Soon-to-be Dr Georgette Tanner - Computational Postdoc
- Ms Rhiannon Barrow - Experimental PhD student
- Ms Muna Al-Jabri - Hybrid PhD student
- Mr Shoaib Ajaib - Computational PhD student
- Ms Nazia Ahmed - Computational PhD student
- Ms Rebecca Prince - Hybrid MRes student And the wonderful Mr Fabio Marcuccio who is an engineering PhD student in Dr Paolo Actis’s lab and is co-supervised by me.
How does HPC help your research?
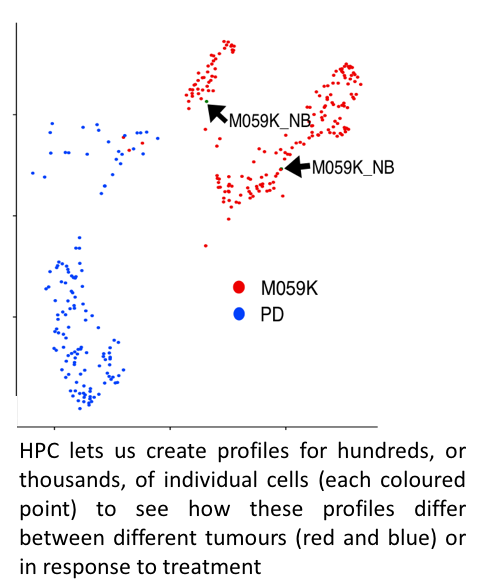
Every cell (well, almost every cell) in a human’s body contains a copy of that persons DNA. DNA is a code of 3 billion ‘letters’. That code is read differently in different cells, making messenger copies of large portions of the DNA, multiple times. This means cells are FULL of data. Add to that: each cell in a tumour is different and we need both levels of information from all of them to understand about how that tumour behaves. Advances in technology are allowing us to mine the information in these cells to an unprecedented level. The HPC is invaluable in then letting us term that data into biological insight.
What is the potential impact of your research?
We hope, one day, we will help to develop better treatments for brain cancer - and extend the predicted survival for a diagnosis of glioblastoma beyond an average of just 14 months.
In your personal opinion what’s the coolest thing about your research?
The fact that we can take a single cell, unable to be seen by the human eye, and produce GB of data on what it is and why to behaves the way it does.
What’s your favourite christmas film?
Bad Santa